Thresholding#
When flattening images and finding grains, TopoStats uses thresholding to separate the background data from the foreground data. This is done by setting a threshold value, and classifying all pixels above this value as foreground, and all pixels below this value as background.
There are several different types of thresholding that can be used, and each has its own advantages and disadvantages.
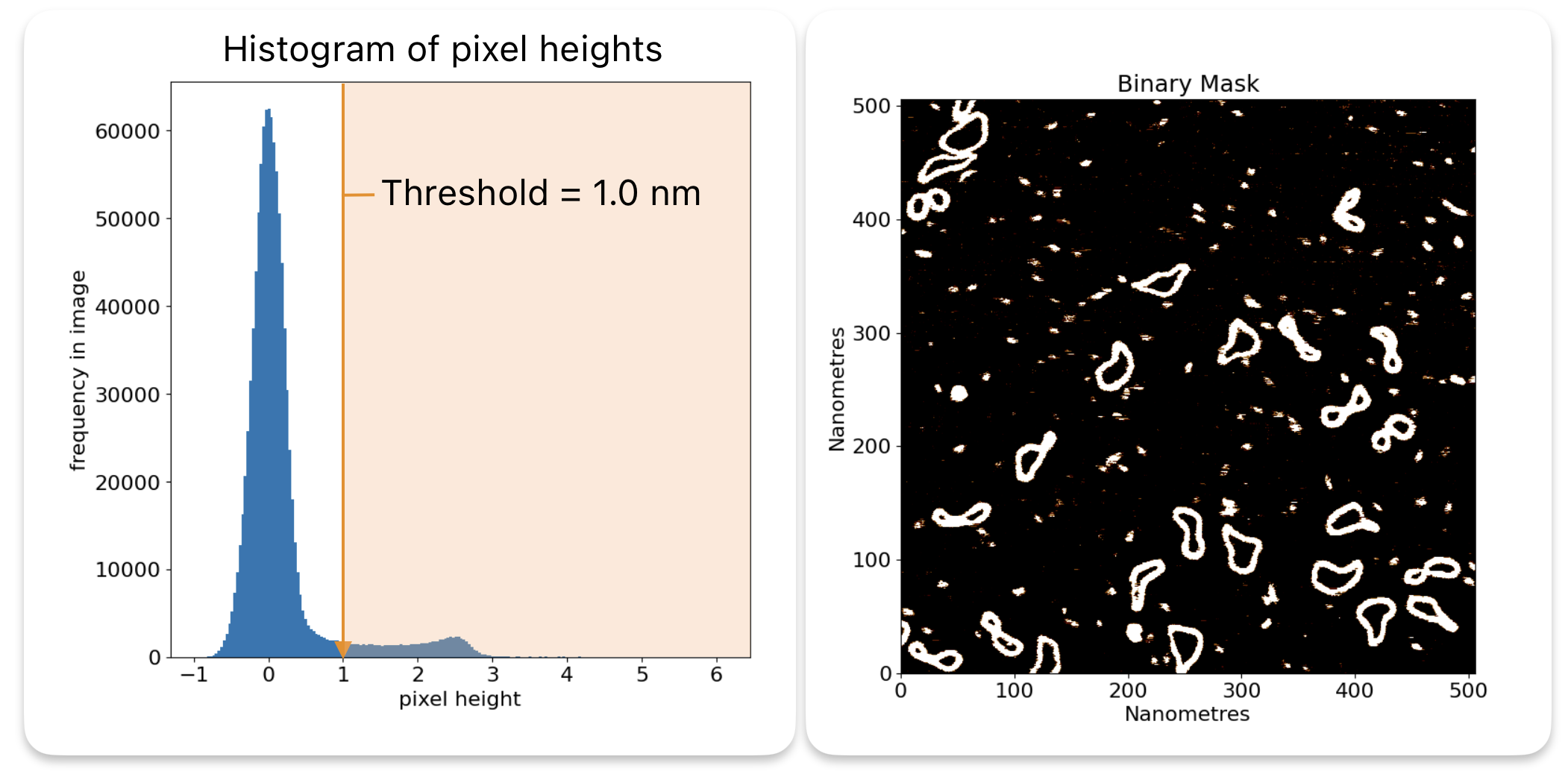
Below is a histogram showing the heights of the pixels in minicircle.spm after flattening. You can see that most of the pixels are at a height of 0nm, which is the background. There is a second peak at 2.5nm which is around the height that we expect DNA to be. The rest of the pixels are noise.
A threshold will select all pixels above a certain height, and ignore the rest, ie the pixels in the orange area.
Note: Thresholding above and below the surface#
TopoStats has the ability to threshold both above the sample surface and below it. This allows finding grains on the surface but also holes in the surface (useful for silicon wafer analysis). This can be configured by setting the corresponding “above” or “below” thresholds in the config file. Eg if you only want to find grains above the surface, only use the “above” threshold options, and vice-versa.
Thresholding types#
Standard deviation thresholding#
Standard deviation thresholding is a simple method of thresholding that uses the standard deviation of the image to determine the threshold value. The threshold value is calculated as:
$$ \text{threshold} = \text{mean} + \text{std_dev} \times \text{factor} $$
Where mean is the mean of the image, std_dev is the standard deviation of the image, and factor is a user-defined
value that determines how many standard deviations above the mean the threshold should be.
This method is useful when you don’t know the exact threshold value you want to use, and when you have a bit of noise in your image.
Otsu thresholding#
Otsu thresholding is an automatic thresholding method that tries to find the threshold value that minimizes the intra-class variance of the foreground and background pixels.
We have added a multiplier to the Otsu thresholding method to allow for a more flexible thresholding method. The threshold value is calculated as:
$$ \text{threshold} = \text{otsu} \times \text{factor} $$
Where otsu is the threshold value calculated by the Otsu method, and factor is a user-defined value that allows you
to adjust the threshold value.
This method is useful when you want to automatically find the threshold value, and when you have a clear binomial distribution of pixels (heights). I.e. separation between the foreground and background pixels in your image with little noise.
Absolute thresholding#
Absolute thresholding is a simple method of thresholding that uses a user-defined threshold value to separate the foreground and background pixels.
This method is useful when you know the exact threshold value you want to use, for example if you know your DNA lies at 2nm above the surface you can set the threshold to 1.5nm to capture the DNA without capturing the background.